This ongoing project extends previous work by Alberts and Odell on actin assembly and Listeria propulsion to explore the functional dynamics of cross-linked actomyosin networks. We have three major goals. The first is to explore in general terms how meso-scale contractile and viscoelastic properties of actomyosin networks emerge from the micro-scale dynamical interactions of actin-filaments, cross-linkers, myosin motors, and factors that regulate their assembly and activities. The second is to couple detailed simulations to our ongoing studies of cortical actomyosin dynamics in early C. elegans embryos (link to polarization page). A third, and longer-term, goal is to infer from detailed simulations of a small patch of cortex the appropriate constitutive laws for a continuum-style model of the whole cell.
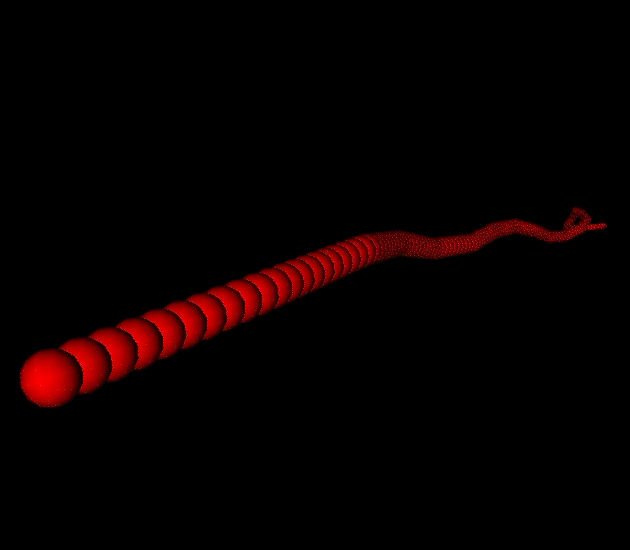
The agents in our simulations are actin filaments (modeled as semi-flexible polymers built from discrete subunits), various cross-linkers, and (not shown in simulations below) myosin mini-filaments. Individual agents move/writhe in response to thermal forces, collide and exchange forces with one another. At every timestep, each cytoskeletal part in the simulation decides what to do next by numerically solving its zero-inertia Newtonian force/torque balance equations of motion. To compute 10 seconds of simulated dynamics for a small patch of meshwork a few microns in diameter takes 10s of hours.Initially, we have focused on calibrating our simulations against empirically measured properties of individual actin filaments and cross-linked filament networks. We have tuned our simulated actin filaments (under thermal forcing) to match experimentally determined values for persistence length, a theoretically derived elastic modulus, and relaxation times when undergoing deformation (click here to check out Jon Albert's demo of how this works called SImFil). We are now calibrating simulations of cross-linked networks against the measured time-dependent rheology of networks assembled from purified components in vitro. See the movies below for some examples.
We have also devised a generic framework for representing different cross-linkers and motors, with variable structures, force-dependent binding kinetics, duty cycles, and preferred angles for filament binding. We are now running simulations simultaneously on the ~450 processors of the CCD's linux cluster to explore how the viscoelastic properties of this complex filament network depend on densities and properties of different cross-linkers and on filament assembly dynamics. In the movies shown below, the cross linker is based on properties of alpha-actinin.
 |
 |
 |
|---|---|---|
A single filament undergoing thermal writhing. |
Forces and Torques acting on two filaments undergoing thermal motion and bound together by linkers. |
Steric Hinderance of filaments |
We are now calibrating the networks behavior to match one-point and two-point microrheology experiments in vitro.
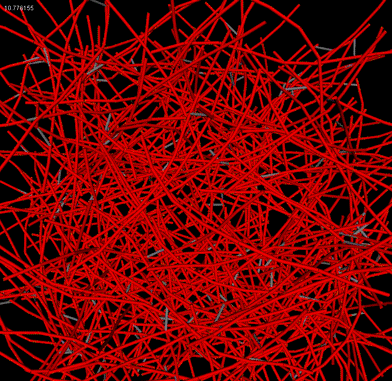 |
 |
 |
|---|---|---|
| Patch of cortex |
Downard flythrough of cortex |
Microrheology of filament network |
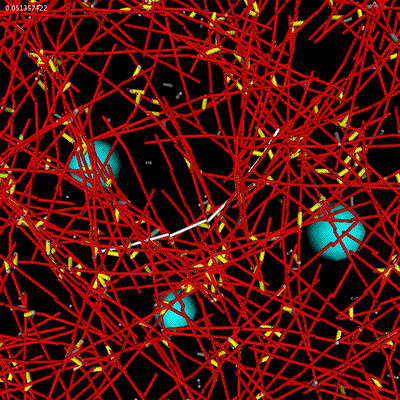 |
||
| Microrheology of filament and linker network. |
The meshwork currently exhibits the qualitative features of bundling for certain types of linkers and gel effects at high linker concentrations.
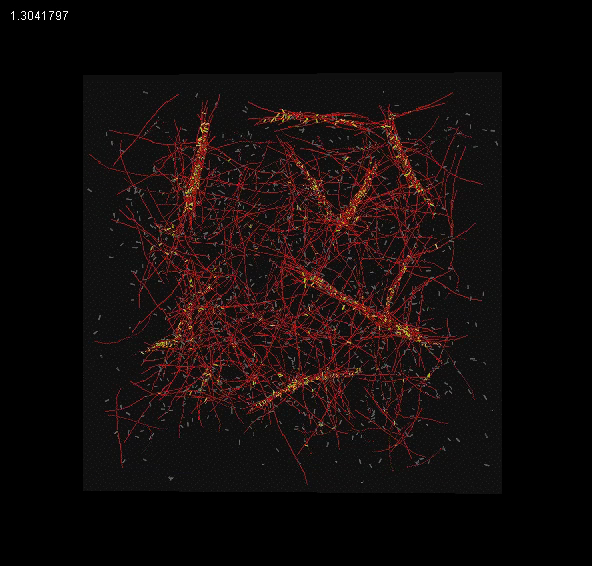 |
 |
|
|---|---|---|
| Filament Bundling |
Gel effects at high linker concentration |
Bead tracking |