ParM is a bacterial actin homolog (Møller-Jensen et al 2002) with unique polymerization dynamics, and unique behaviors in vivo. ParM filaments exhibit equal end dynamics, unlike actin filaments, and ends can transition from stable elongation to rapid disassembly, much like microtubules (Garner et al 2004). As with microtubules, end stability is presumably modulated by a “cap condition”, i.e. the hydrolysis state of some terminal number of ParM monomers determines whether a particular filament end can stochastically elongate. These unique biochemical behaviors likely facilitate the in vivo mechanical role of this filament: the faithful segregation to daughter bacteria of the plasmids that encode the ParM gene.
The rod-shaped bacteria (such as E. coli) in which ParM forms this prokaryotic mitotic spindle are small, with typical length and diameter of 3 µm and 1 µm, respectively. At ~12 µM, there is only enough ParM monomer to span the bacteria several times. Thus, plasmid segregation proceeds from a small filament population, and the behavior of each individual filament is potentially important. To appropriately model this segregation, we have constructed a 3-dimensional force-based simulation in which each ParM filament is tracked, monomer-by-monomer, from nucleation to hydrolysis to disassembly. In vivo, and in the full in silico model (presented elsewhere), a filament’s elongation may be further modified by interactions with the bacterial crucible, other filaments, and most importantly the plasmids, which likely alter polymerization kinetics. We are working to determine the likely nature of the polymerization cap condition by comparison of in vitro results (e.g. bulk filament assays and total internal reflection fluorescence (TIRF) observations of individual ParM filaments) with equivalent in silico experiments.
Previous in vitro observations define a concentration trajectory and length distribution, and suggest that a polymerization “cap condition” may govern filament stability. We test the nature of this cap condition, with a stochastic computer model, under both independent and cooperative hydrolysis mechanisms. With that independent hydrolysis with a cap size of three ATP monomers can best explain our experimental observations.
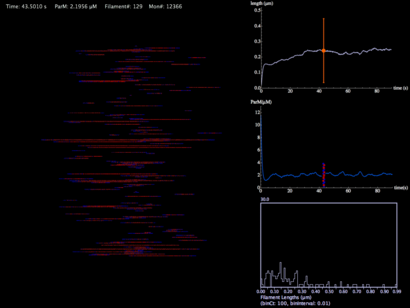
This movie is a force-less simulation of a ParM filament population from nucleation, to hydrolysis, to depolymerization. The monomer concentration is initially 12 µM in a pill-shaped volume 3 microns in length and 1 micron in diameter (a typical bacterial volume). Graphs show average filament length with standard deviation, ParM concentrations, and a histogram distribution of filament lengths.
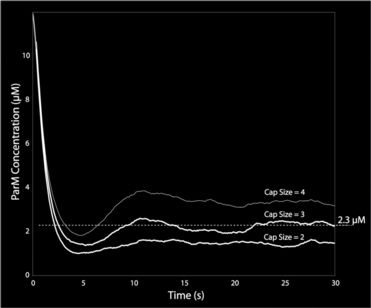
This graph shows ParM monomer concentration for independent hydrolysis (stochastic hydrolysis that is independent of neighbor monomer states) and three different cap-size assumptions in the model. Only with a cap-size of 3 monomers (i.e. there must be three terminal ATP monomers for continued polymerization) do we get the right steady-state ParM concentration of 2.3 µM.
With all the crucial biochemical rates in hand, we can now reasonably begin to approach actual plasmid segregation with a force-based model. Filament flexibility if modeled by the PAIRS method we developed, which ensures realistic deflections, relaxation time-constants, and thermal motions of the ParM filaments. We are using in vivo observations of plasmid diffusivity (Campbell et al, 2008) to estimate the effective viscosity for filaments (which is likely body-axis dependent). Still, the nature of many mechanistic interactions and environmental parameters remain uncertain, and we are in the process of exploring the segregation parameter and mechanism space.

|
|
An apparently successful plasmid segregation. I like this movie --it looks good to my mind's eye-- but it suffers from improper filament flexibility, and uncertain filament-filament side-binding. |
The idea here is to reproduce the observed behavior with a minimum of ad-hoc assumptions and mechanisms (though since this model is very detailed oriented, we have to assume the nature of some unknown interactions). The first part of this story, the biochemical simulations that inform us about the nature of the cap condition, is nearing publication.